LigandScout supports data export in several chemical and
graphical file formats. Use the command (CTRL+SHIFT+S) in the menu
>
which enables you to
save your macromolecule, ligand modifications, and pharmacophore models.
A session can be stored in the menu
>
>
.
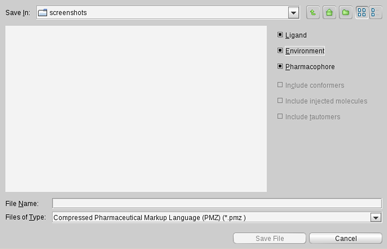
Depending on the selected file format and content, the
Save File
dialog provides
advanced export settings
.
The following table
gives you a summary of the supported export file formats together with which data
of your work that can be exported in a particular format.
Each format facilitates the export of certain information:
molecules (M), ligand environment (E), pharmacophore models (P), and
the current content of either 3D View or 2D View (V).
LigandScout has its own file format, called PML. The
advantage of this format is that the user can select the available
export content individually or in combination (ligand, environment,
pharmacophore, conformers, injected molecules, and tautomers).
You can save your content as PDB or ENT file format if you want to
export the ligand, environment and include hydrogen information.
The SMILES string of molecules can also be exported as SMI files. You can include
in the files the canonical order of ligand atoms and/or stereo chemical properties.
Single molecules can be stored in mol files, as 2D or 3D data. Additionally,
stereo-chemical properties can be saved optionally.
The storage of a molecule
library in LDB, MOL(2) and SD(F)
files can be customized. You can export a
Complete library
or a part of it.
A subset of a library can be chosen by
filtering your library
according to a property (e.g. by an occurring name), marking molecules or
by visible molecules only.
If you want to export the properties of your library table, choose the
Write properties
option.
The MOL2 and SD(F) file formats
offer you additionally the possibility to
Write all conformations
or
Write 2D/3D coordinates
of the (selected)
molecules in the library.
If you want to export currently
displayed content as an image file (e.g. gif, svg, etc.)
you can choose between the 2D or 3D View. The 2D content can be saved as it
is by the
Keep current layout
or you can automatically
store it as
horizontal/vertical layout
. After
a screening run, you can export the
ROC plot
as image.
The image size can be set in both 2D and 3D.