Why Use Structure-Based Pharmacophores?
Pharmacophores represent chemical functions,
valid not only for the currently bound, but also unknown molecules.
Due to its simplicity, this method is computationally very efficient
and is subsequently exceptionally well suited for virtual screening
of large compound libraries. Pharmacophore modeling is universal,
comprehensive and editable: Selectivity-tuning by adding or
omitting feature constraints provides a plenitude of manipulation
options.
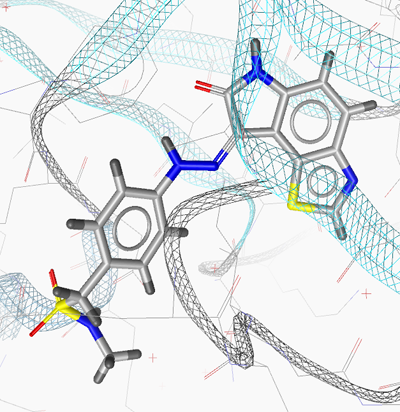
The following figures show the active site of CDK2 with
ligand LS2 (PDB 1KE6) and the respective pharmacophore model
generated by LigandScout. The program automatically searches for
interactions between ligands and the macromolecule.
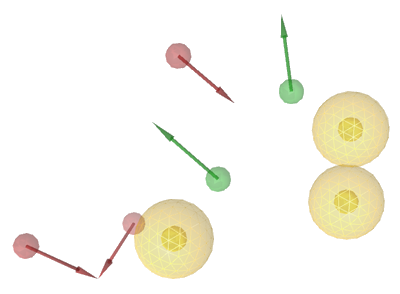
In this example, LigandScout suggests an eight feature
pharmacophore model consisting of three hydrophobic features
(yellow spheres), two hydrogen bond donor features (green
vectors) and three hydrogen bond acceptors (red vectors). This
pharmacophore model represents a highly potent filter for in-silico
screening of new CDK2 ligands.