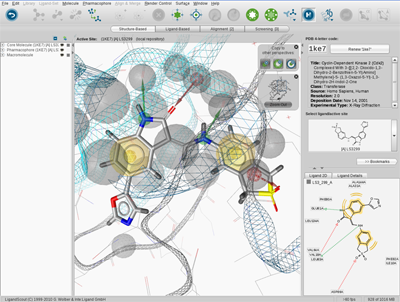
LigandScout Graphical User Interface
LigandScout uses an intuitive graphical user
interface (GUI) for sophisticated pharmacophore generation and data analysis.
It provides five perspectives, which have different functionalities:
-
Structure-Based Modeling Perspective
Analysis of ligand-macromolecule interactions.
Viewing and calculating properties of molecule libraries in the PDB View.
Managing binding sites in the Bookmark View.
-
-
-
The
Figure 2.2, “Graphical user interface”
shows the Structure-Based Modeling Perspective of LigandScout.
LigandScout contains a menu, toolbar, five perspectives and control panels
(in this case, the PDB Panel is shown). Furthermore, LigandScout provides modules such as interactive
2D and 3D visualization areas,
Library View
and
Hierarchy View
,
which help to orientate and navigate through your data. The visibility of the control panels,
2D View
and
Hierarchy View
can be toggled in the
menu.
LigandScout provides universal
Undo
and
Redo
functionality that lets you go back and forth in history.