Why Use Ligand-Based Pharmacophores?
The ligand-based
strategy deals with the absence of the macromolecule structure by
deriving pharmacophore models from a set of ligands.
This approach considers the conformational flexibility of the ligands.
Similar small molecules provide similar biological activity.
Thus, this approach searches for a common feature pattern that is shared
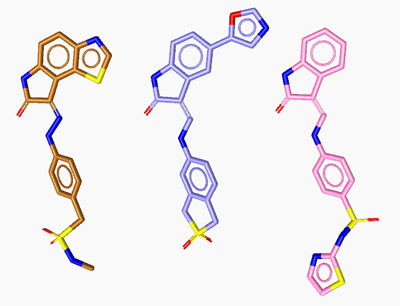
in an active ligand-set. The following figures show the ligands
(LS2, LS3 and LS4) of 1ke6, 1ke7 and 1ke8 which bind to CDK2.
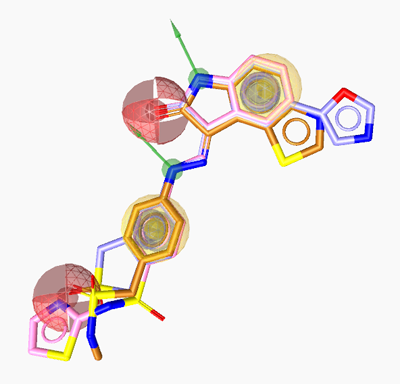
LigandScout automatically creates the respective pharmacophore to these ligands.
In this case, LigandScout generates a ligand-based pharmacophore
which is composed of two hydrophobic features (yellow spheres),
two aromatic rings (grey elements),
two hydrogen bond donor features (green vectors) and two hydrogen
bond acceptors (red vectors).