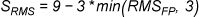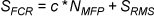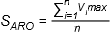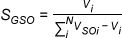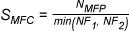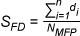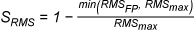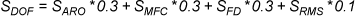LigandScout provides several scoring functions for ranking intermediate solutions during the
ligand-based pharmacophore generation. All scoring functions were
developed to deliver robust results for several investigated targets. However, different scoring functions may
deliver different results, depending on whether shape (i.e., molecular volume) or chemical functionality is
considered to be more relevant for the assumed binding mode. Available scoring functions comprise
Pharmacophore-Fit (only taking into account chemical feature overlap), Atom Overlap
(“
Fast Shape
”), Gaussian
Shape Similarity, and a combination of Pharmacophore-Fit and Atom Overlap.
These scoring functions are described in detail below.
Simple Geometric Scoring Function (“
Pharmacophore-Fit
”)
Atom Sphere Overlap (“
Fast Shape
”)
Gaussian Shape Similarity
Pharmacophore-Fit and Atom Overlap