
LigandScout
User Friendly Advanced Molecular Design
The LigandScout software suite comprises the most user friendly molecular design
tools available to chemists and modelers worldwide. The platform seamlessly
integrates computational technology for designing, filtering, searching and
prioritizing molecules for synthesis and biological assessment.
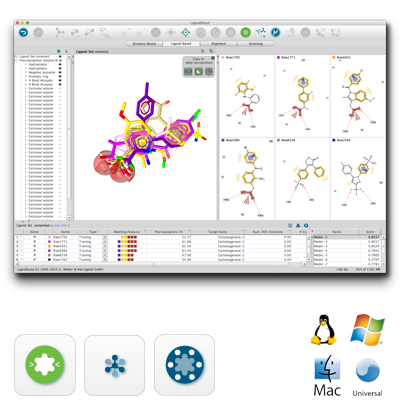
Our advanced algorithms for 3D pharmacophore modeling, virtual screening,
activity profiling and virtual library design create an inspiring and productive
experience for multidisciplinary teams. By providing excellent prediction quality
with unprecedented speed, LigandScout successfully addresses challenges in
interactive molecular design.
The algorithms are scientifically validated [1-4] and based on multiple years of experience in modeling software design, while the application corresponds to state-of-the-art information technology.
The full-featured 3D graphical user interface with multiple undo-levels makes the molecular design process efficient and transparent. Cloud computing is available from the GUI via the LigandScout Remote [5] option.
LigandScout runs on all common operating systems, and is available in three different options: LigandScout Essential, LigandScout Advanced, and LigandScout Expert. Additionally, we offer the entire LigandScout functionality and some additional tools in the Inte:Ligand LigandScout Knime Extensions package.
Free evaluation: You can download a fully functional version and test it for
one month. To obtain the download links register here or contact us at support@inteligand.com
Publications describing usage of LigandScout on Google Scholar
For a recent list of papers click here
- Wolber, G.; Langer, T.; LigandScout: 3-D Pharmacophores Derived from Protein-Bound Ligands and Their Use as Virtual Screening Filters.
J. Chem. Inf. Model; 2005; 45(1); 160-169.
DOI: 10.1021/ci049885e (request reprint) - Wolber, G.; Dornhofer, A. A.; Langer, T.; Efficient overlay of small organic molecules using 3D pharmacophores
J. Comput. Aided Mol. Des.; 2007; 20(12); 773-788.
DOI: 10.1007/s10822-006-9078-7 - Wolber, G.; Kirchmair H.; Langer, T.; Structure-Based 3D Pharmacophores: An Alternative to Docking?
Plenary talk at the 7th International Conference on Chemical Structures in Noordwijkerhout, 2005
Presentation as PDF file [2.8 MB] - Wolber G., Dornhofer A. A., Langer T.; Efficient Overlay of Molecular 3D Pharmacophores. Oral presentation at the spring ACS meeting in Atlanta, GA, USA, 2006
Presentation as PDF file [8.8 MB] - Kainrad T., Hunold S., Seidel T., Langer T.; LigandScout Remote: A New User-Friendly Interface for HPC and Cloud Resources
J. Chem. Inf. Model; 2018.
DOI: 10.1021/acs.jcim.8b00716
