
LigandScout
Advanced Molecular Design
Structure-based 3D Molecular Design
-
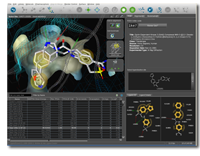 Automatic interpretation of PDB ligands using geometry, dictionaries and rules
Automatic interpretation of PDB ligands using geometry, dictionaries and rules
- State-of-the-art user interface with advanced 3D graphics and undo-function
- 2D view and hierarchical view directly linked to 3D interface
- Fast alignment of molecules in their bio-active conformation to other molecules and 3D pharmacophores from several ligands and/or pharmacophores, shared feature pharmacophores can be derived to understand and model the relevant mode of action
- Advanced handling of co-factors, ions, water molecules and covalently bound ligand
- Extensive parameter control for more experienced users
- Advanced PDB ligand perception and easy manual correction while modeling in the active site
- Ability to treat co-factors and water molecules as part of the ligand or part of the macromolecule
- Intuitive pharmacophore-based alignment of molecules
- Support for most common file formats
- Sophisticated file and repository management of edited and stored binding sites, molecules and pharmacophores
- Smart enumeration of tautomers
Accurate and Fast Virtual Screening
- High performance accurate virtual screening with automated analysis of screening performance using ROC curves and enrichment factor calculations
- Simple workflows for boolean combination of target and anti-target pharmacophore models
- Easy to use implementation of AutoDock 4.2 and AutoDock Vina
- Accurate implementation of the MMFF94 force field for generating high quality geometries
Ligand-based Pharmacophore Design
- Ligand-based pharmacophore modeling, including automatic classification of chemical features, feature weights and generation of exclusion volume spheres
- Automated training set selection by pharmacophore-based cluster analysis
System Requirements
- Intel i5 processor or equivalent, at least 2GB of RAM per core (min 4GB recommended)
- One of the following 64bit operating systems: Windows, Linux, macOS or MacOS X
- Hardware-accelerated 3D graphics card (NVIDIA recommended) with OpenGL 1.2 support

|
Register and get LigandScout now! |
