|
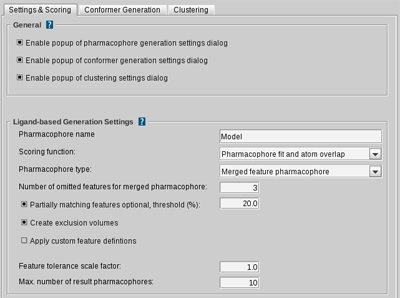
Pharmacophore name
|
Sets the default name (prefix) for the resulting pharmacophores. (default: Model)
|
|
Scoring function
|
During the ligand-based pharmacophore generation all intermediate alignment results
are scored according to the selected scoring function. The
Pharmacophore-Fit
scoring
function only considers pharmacophoric features and the feature RMS deviation.
The
Relative Pharmacophore-Fit
scores the number of
matching pharmacophore features and the RMSD of the pharmacophore
alignment normalized to [0..1].
If you want to consider steric properties, you can choose the
atom sphere overlap (fast shape)
or
Gaussian shape similarity
scoring functions.
The first implements a fast
atom sphere counting algorithm to provide an overlap score. The latter,
uses Gaussian functions to approximate atom spheres. By doing so the overlap can be
calculated analytically. (default: Pharmacophore-Fit and atom overlap)
|
|
Pharmacophore type
|
Two different types of pharmacophoric models can be derived due to
different generation processes. The ligand-based pharmacophore generation
is a pair-wise process. In each step one pharmacophore for two molecules is
drained. This we can do by either taking all common features
(Shared Feature Pharamacophore) or by taking all features (Merged Feature
Pharmacophore). In the latter case each feature is scored and those are
removed that do not match all input molecules (see number of omitted features
for merged pharmacophore). The
shared feature pharmacophore
creates a
pharmacophore that represents all common features of a ligand-set.
If you want to take all features into account and assemble them into one pharmacophore,
then the
merged feature pharmacophore
is your right choice.
(default: Merged feature pharmcophore)
|
|
Number of omitted features for merged pharmacophore
|
This setting specifies the number of features that need not
match all input molecules if
Merged feature pharmacophore
type is turned on. (default: 3)
|
|
Partially matching features optional, threshold (%)
|
If this threshold is exceeded, e.g. more than 20% of the molecules do not match a feature,
the feature is declared as optional, otherwise it is handled like a normal feature.
(default: 20)
|
|
Create exclusion volumes
|
Excluded Volumes are generated.
(default: enabled)
|
|
Apply custom feature settings
|
Enables customized feature definitions.
(default: disabled)
|
|
Feature tolerance scale factor
|
This value is multiplied to the pre-defined feature tolerances. Lowering this scale factor
would end up in more restrictive pharmacophoric models.
(default: 1.0)
|
|
Max. number of resulting pharmacophores
|
The maximum number of pharmacophores to be generated.
(default: 10)
|