2.1.2. Fragments view
ilib diverse uses a huge collection of fragments for the generation of compound libraries. Every fragment is assigned to a fragment group as shown in the screenshot below.
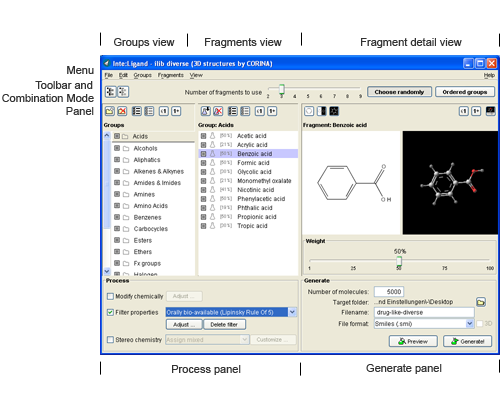
Adding fragments
Click on the
![]() icon or use the
import > fragment set and flasks command from the
file menu to import new
fragments (from SMILES files or MDL MOL files) to the current group.
icon or use the
import > fragment set and flasks command from the
file menu to import new
fragments (from SMILES files or MDL MOL files) to the current group.
Deleting fragments
Delete one or more fragments from the currently selected group by clicking on the
![]() icon or by pressing
SHIFT+DEL. Multiple fragments can be selected as described below.
icon or by pressing
SHIFT+DEL. Multiple fragments can be selected as described below.
Selecting/deselecting fragments
Click on the checkbox of each fragment in the fragments view to select/deselect fragments.
Hold down the SHIFT key for selecting a range of fragments. In order to select or deselect several single fragments hold down the CTRL key.
Select/deselect all fragments of the current group by clicking on the icons
![]() and
and
![]() . Alternatively, you may use
the use all fragments of the selected group and the do
not use any fragment of the selected group commands from the
fragments menu.
. Alternatively, you may use
the use all fragments of the selected group and the do
not use any fragment of the selected group commands from the
fragments menu.
Adjusting the occurrence of fragments in generated molecules
Use the icons
![]() and
and
![]() to control the frequency of occurrence
of the selected fragments in generated molecules.
to control the frequency of occurrence
of the selected fragments in generated molecules.
By clicking on
![]() , the current
fragment will be used at least once for library molecule generation.
Clicking on
, the current
fragment will be used at least once for library molecule generation.
Clicking on
![]() will result in integrating
the selected fragment at most once for building the library molecule.
will result in integrating
the selected fragment at most once for building the library molecule.
When activating both modes
(![]() and
and
![]() ) the selected fragment(s) are used
for the generation of all molecules.
Alternatively, you may use the use selected fragment at least once and
the use selected fragment at most once commands from the
fragments menu.
) the selected fragment(s) are used
for the generation of all molecules.
Alternatively, you may use the use selected fragment at least once and
the use selected fragment at most once commands from the
fragments menu.
Renaming a group
Rename the currently selected group using the rename selected item command from the edit menu. Alternatively, you may rename a group by double-clicking or by pressing the F2 key after selection.
