Example 2: Generation of a Focused Virtual Library of 4-Aminoquinoline Derivatives as NMDA-NR1/2B Antagonists
Introduction and Pharmacological Background
Excessive activation of N-methyl-D-aspartate (NMDA) receptors and resulting calcium overload of neurons is thought to be a key contributor to neuronal cell death following acute cerebral ischaemia. In this context, NMDA receptor antagonists have been demonstrated to be potent neuroprotective agents in animal models of focal cerebral ischemia. Therefore, selective NR1/2B subtype antagonists are considered as potentially attractive drugs for the treatment of neurodegenerative disorders such as stroke, brain trauma, pain, and Parkinson's disease. [1]
Template Molecules for Focused Library Generation
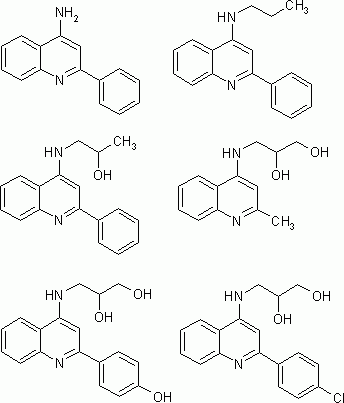
Recently, 4-aminoquinolines have been discovered as a novel class of NR1/2B subtype selective NMDA receptor antagonists. [2] In general, compounds of this class consist of quinoline as the core structure with substituents at the positions 2 and 4. At position 2, a hydrophobic residue is substitued, while at position 4, a more hydrophilic residue consisting of a short chain amine with none, one, or more hydroxy groups contributes to the activity of the compound.
Figure 1: Structure templates of 4-aminoquinoline NMDA-NR1/2B antagonist compounds (examples)
Library Generation with ilib diverse
Based on the general structure of this NMDA antagonist class, we define three flasks for the library generation process. It is more practicable to generate a molecule from one end to the other, so we do not start the library generation at the structure core. All fragments are given a weight of 100%.
Figure 2: Division of 4-aminoquinoline NMDA-NR1/2B antagonist compounds into three building blocks used for flask definition in ilib diverse
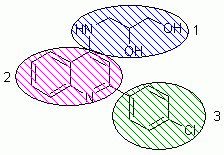
Flask 1 (blue): We start at the hydrophilic end of our molecules. Flask 1 contains Amine (from the Fx groups), Colamine, and Propylamine (from the Amines group). Based on the literature [2], we construct four new fragments: 2-Hydroxylpropylamine, 3-Hydroxylpropylamine, 2,3-Dihydroxypropylamine, and Aminoacetone. These fragments are saved as *.mol files, imported into the Amines group and added to Flask 1. The reactivity of all fragments is set to 10 at the nitrogen and zero at all other atoms to assure substitution at the correct position. "Reset reactivity after first substitution" is highlighted with each nitrogen.
Flask 2 (magenta): This group bears the core structure, Quinoline. The fragment is added from the Heterocycles, bicyclic group. Reactivity is set to 10 at position 4, where the first substitution with the fragments from Flask 1 should take place, "Reset reactivity after first substitution" is highlighted. At position 2, reactivity is set to 4 to allow the second substitution with the fragments from Flask 3. All other position are set to a reactivity of -10 wherever possible.
Flask 3 (green): Hydrophobic moieties and substitued benzene rings are added to Flask 3. We select Methane, Ethane, and Propane from the Aliphatics group. Furthermore, Anisole, Benzene, Chlorobenzene, Phenol, and Toluene from the Benzenes group join Flask 3. The reactivity of Propane is set to 10 at position 2 to gain an isopropyl substitution. All reactivities of substitued benzenes are set to 10 at position 4 to ensure p-substitution.
Recommended Filter Settings
Application of the ‘Lipinski rule of five’ filter ensure high drug-likeness and oral bioavailability of the generated compounds. Stereo chemistry setting is defined as by default (assign mixed stereo chemistry).
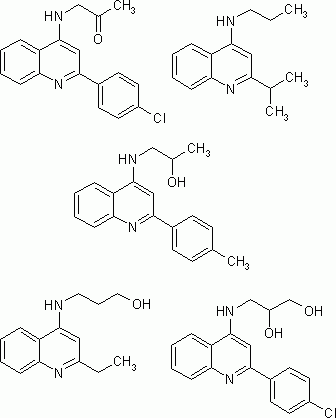
This generation process results in a drug-like library of compounds that share the same shape and the same functions as known NMDA antagonist molecules. They are therefore promising candidates for synthesis and biological testing. On standard hardware, time consumption for each generated compound came up to 33ms. Out of the generation process, one of the structures did not match the filter requirements - the desired estimated log P - and was therefore rejected. The Report Section provides information on the generation process as well as on the estimated log P and molecular weight distribution. Examples of generated compounds are given in Figure 3.
Figure 3: Examples of generated structures showing high NMDA antagonist analogy
References
[1] Alanine, A. et al. 1-Benzyloxy-4,5-dihydro-1H-imidazol-2-yl amines,
a Novel Class of NR1/2B Subtype Selective NMDA Receptor Antagonists.
Bioorganic & Medicinal Chemistry Letters, 2003, 13, 3155-9
[2] Pinard, E. et al. 4-Aminoquinolines as a Novel Class of NR1/2B
Subtype Selective NMDA Receptor Antagonists. Bioorganic & Medicinal
Chemistry Letters, 2002, 12, 2615-9
To use the fragment set and flask settings described here, import the file nmda-ant.ifs from the examples directory.
