Example 4: Potential Inhibitors for Human Coagulation Factor Xa
Introduction and Pharmacological Background
The prevention of blood coagulation is a major topic considering serious physiological reactions of undesired blood clotting that can lead to myocardial infarction, stroke, deep vein thrombosis and pulmonary embolism. Factor Xa is a trypsin-like serine protease that converts the prothrombin zymogen to its active form thrombin and initiates the final stage of the blood coagulation pathway. Thrombin catalyzes enzymatic reactions that result in the formation of fibrin, the important binding part for blood clotting. Compared to thrombin, factor Xa acts on an earlier level than thrombin. The mechanism of amplification in the blood coagulation cascade causes a small amount of factor Xa to produce a large amount of thrombin. Hence, the direct inhibition factor Xa is considered as more favorable regarding the safety/efficacy ratio compared to other anticoagulants. Other anticoagulant substance classes like vitamin K antagonists or thrombin inhibitors are considered to have a higher bleeding risk than factor Xa inhibitors. Therefore, inhibition of factor Xa has appeared as an attractive target for the development of new anticoagulants. [1]
Template Molecules for Focused Library Generation
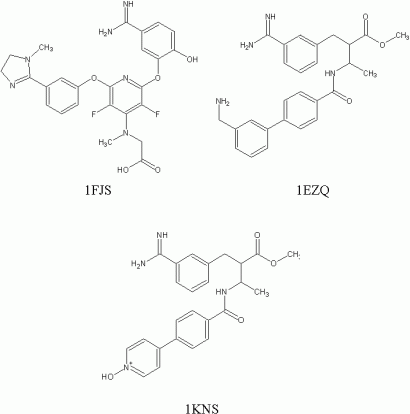
Beside the numerous recently published work dealing with the rational design of factor Xa inhibitors, several entries of human factor Xa inhibitors are deposited in the Protein Data Bank. [2] Due to reasons of straightforwardness, three of the PDB Entries (1KSN, 1EZQ, and 1FJS) are shown and used to guide our virtual library generation process. The ligands show affinity data (Ki values) in the sub-nanomolar range against human factor Xa. The structural templates of the three ligands from the corresponding PDB entries are presented in Figure 1. [3,4,5]
Figure 1: Structure templates of three highly active factor Xa inhibitors (examples)
Library Generation with ilib diverse
Based on the structure of the factor Xa inhibitor templates we defined seven flasks for the library generation process. It is more practicable to generate a molecule from one end to the other, so we do not start the library generation at the structure core.
Figure 2: Division of factor Xa inhibitors into building blocks used for flask definition in ilib diverse
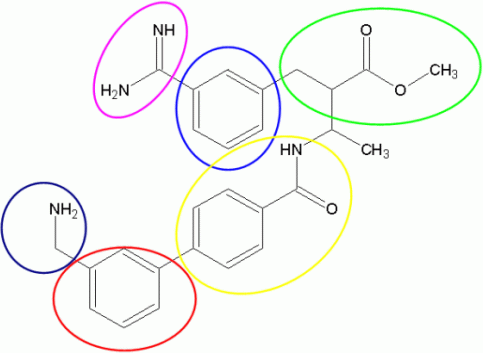
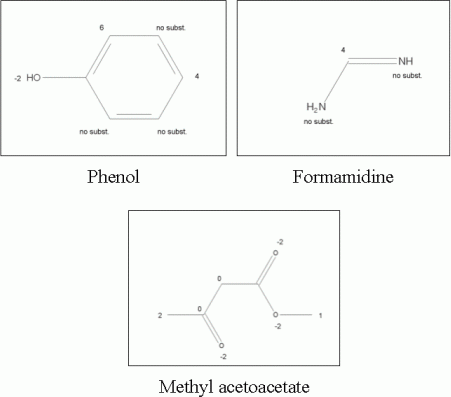
Flask 1 (blue): To start at a hydrophobic part of the molecules, Benzene and Phenol (Benzenes group), Naphthalene (Carbocycles group) and several selected heterocycles (Indole, Thiophene, Coumaron, Benzisoxazole) were included in flask 1. The reactivity was increased to value 6 and 4 in position 1 and 3 of the benzene and position 2 and 4 of the phenol moiety. Further, the substitution was omitted on the rest of the cyclic carbon atoms to ensure the desired structural pattern. The reactivity of the other fragments was assigned is a similar mode. The modified reactivity settings are shown in Figure 3. A weight of 100% was set on the Benzene fragment and weight of 25% was set to all other fragments of flask 1.
Flask 2 (magenta):The Ester and Alcohol groups form the second flask. We restricted the included esters to Ethyl acetate, Methyl acetate and Methyl acetoacetate. Following modified reactivity settings were applied: The terminal carbon atoms were set to higher activity value, e.g. 3 and the button "Reset reactivity after first substitution" is highlighted for each atom with increased activity. Hence, the multiple substitution on one carbon atom is reduced significantly. An example is presented in Figure 3.
Flask 3 (green): The Formamidine moiety is selected to substitute Flask 1 in one of the earlier defined positions. A modification of the reactivity parameter was applied and is presented in Figure 3. Furthermore, the "Reset reactivity after first substitution" button was selected to omit a multiple substitution of the carbon atom of the Formamidine fragment.
Flask 4 (yellow): All selected fragments for flask 4 are derived from the Carbocycles group (Naphthalene, Indene, Cyclohexene), the bicyclic Heterocycles group (Benzisoxazole, Coumaron, Indole, Isoquinoline) and the Aliphatics group (Ethane, Methane). The modified reactivity values were kept to lead to chain-shaped molecules and prevent a high degree of substitutions on single fragment parts. The "Reset reactivity after first substitution" button was selected for this reason as well.
Flask 5 (red): The next hydrophobic part of the molecules was presented by a selection of different structural moieties in flask 5. Beside the Benzene fragment, other entries from the Carbocycles group (Naphthalene, Indene, Cyclohexene), the monocyclic Heterocycles group (1,3-Thiazole, Furan, Imidazole, Indolizine, Isothiazole, Oxazole, Pyrazole, Pyridine, Pyrimidine, Pyrrole, Pyrrolidine, and Thiophene), Aliphatics group (Ethane, Methane), and the bicyclic Heterocycles group(Benzisoxazole, Coumaron, Indole, Isoquinoline) were chosen. Another added fragment was the Sulfonamide, a substructure that is found in several factor Xa inhibitors. [6]
Flask 6 (dark blue): The settings for flask 6 were selected to extend the length of the chain within the direction of the fragments that were used on flask 4 and 5. The monocyclic and bicyclic Heterocycles groups (1,3-Thiazole, Furan, Imidazole, Indolizine, Isothiazole, Oxazole, Pyrazole, Pyridine, Pyrimidine, Pyrrole, Pyrrolidine, Thiophene, Benzisoxazole, Coumaron, Indole, and Isoquinoline) were chosen beside the Amines group containing Butlyamine, Colamine, Diethylamine, Dimethylamine, Ethylamine, Methylamine, Morpholine, Piperazine and Piperidine. This flask was selected in order to generated molecules that can also include structures of the Amines group.
Flask 7 (dark blue): The final building block for the focused library was an addition of aliphatic moieties, like methane and ethane. The reactivity values are kept the same as in flask 4.
The fragment selection for Flask 4-7 were considered to find new scaffolds with similar chemical and spatial demands as active factor Xa inhibitors. The generated molecules were desired to be not too similar in their topological properties to stretch the chemical and structural variability of the compounds in the generated focused virtual library. The weight settings were kept as defined in the default parameter of every single compound.
Figure3: Examples of reactivity settings for fragments used in the generation process
Recommended Filter Settings
Application of the ‘Lipinski rule of five’ filter ensures high drug-likeness and oral bioavailability of the generated compounds. The Stereo chemistry setting was defined to eliminate all compound with a stereo center (R/S). This restriction was used to show that a library can be generated without a single chiral molecule in order to find compounds that are readily to synthesize.
This generation process results in a drug-like library of compounds that share the similar shape and functions as known factor Xa inhibitors. On standard hardware, time consumption for each generated compound came up to 137ms. The time per included library molecule (excl. filtered compounds) came up to 2525ms. This time expense corresponds to the highly restrictive filter settings in order to find only achiral substances. The library generation efficiency can be enhanced by the selection of different fragments and/or fragment weights, which do not contradict filter definitions.
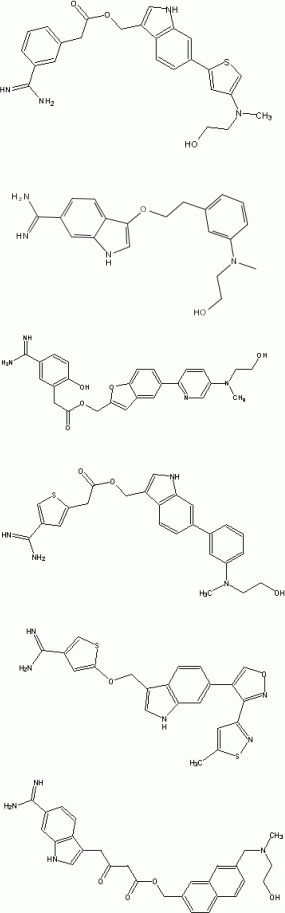
Examples of generated compounds are given in Figure 4. The Report Section provides information on the generation process as well as on the estimated log P and molecular weight distribution.
Figure 4: Examples of generated structures with new new scaffolds and similar chemical and spatial demands as active factor Xa inhibitors.
References
[1] Ostrovsky, D. et al. Analysis of Activity for Factor Xa Inhibitors
Based on Monte Carlo Simulations. J. Med. Chem. 2003, 46, 5691-5699.
[2] Protein Data Bank:
www.rcsb.org/pdb/
[3] Guertin, K. R. et al. Optimization of the beta-Aminoester Class of
Factor Xa Inhibitors. Part 2: Identification of FXV673 as a Potent and
Selective Inhibitor with Excellent in Vivo Anticoalulant Activity.
Bioorg. Med. Chem. Lett. 2002, 12, 1671-1674.
[4] Maignan, S. et al. Crystal Structure of Human Factor Xa Complexed
with Potent Inhibitor. J. Med. Chem. 2000, 43, 3226-3232.
[5] Adler, M. et al. Preperation, Characterization, and the Crystal
Structure of the Inhibitor ZK-807834 (CI-1031) Complexed with Factor
Xa. Biochem. 2000, 39, 12534-12542.
[6] Pruitt, J. R. et al. Discovery of
1-(2-Aminomethylphenyl)-3-trifluoromethyl-N-[3-fluoro-2'-
(aminosulfonyl)[1,1'-biphenyl)]-4-yl]-1H-pyrazole-5-carboxyamide
(DPC602), a Potent Selective, and Orally Bioavailable Factor Xa Inhibitor. J.
Med. Chem. 46, 5298-5315.
To use the fragment set and flask settings described here, import the file fxa-i.ifs from the Examples directory of your CD.
